



Next: Maximum number/length of sequences
Up: usersguide
Previous: redo_mlrgd and redo_plots
Contents
Run time
The conservation programme mlrgd is very quick. On my LINUX PC
with a 2.6 GHz Pentium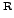 4 processor) it takes
4 processor) it takes
 20-40 s (depending on the parameter options) to run mlrgd
on the Hepatitis B example alignment. Much more time-consuming is the
initial alignment (with code2aln) and the phylogenetic tree
calculation with dnaml (dnapars is much quicker and is
recommended for alignments of more than about 15 sequences). The time
requirements of code2aln and dnaml are very dependent on
the number of sequences. The scripts redo_mlrgd and redo_plots allow one to rerun the conservation programme and redo
the plots with different parameters without having to rerun code2aln and dnaml.
20-40 s (depending on the parameter options) to run mlrgd
on the Hepatitis B example alignment. Much more time-consuming is the
initial alignment (with code2aln) and the phylogenetic tree
calculation with dnaml (dnapars is much quicker and is
recommended for alignments of more than about 15 sequences). The time
requirements of code2aln and dnaml are very dependent on
the number of sequences. The scripts redo_mlrgd and redo_plots allow one to rerun the conservation programme and redo
the plots with different parameters without having to rerun code2aln and dnaml.
aef
2007-12-10